Cusabio Microbiology Recombinants
Genetic information in microbes
The genetic information in bacteria and many viruses are encoded in DNA, but some viruses use RNA. Genome replication is essential for the inheritance of genetically determined traits. Gene expression generally involves the transcription of DNA into messenger RNA and the translation of mRNA into protein.
Genome organization
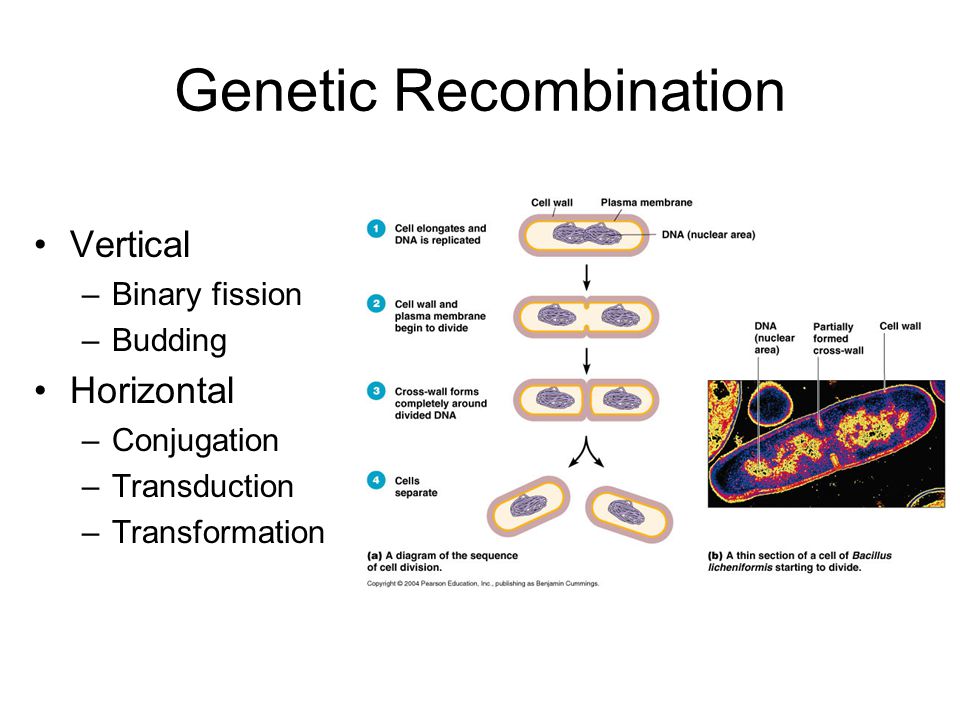
The bacterial chromosome is a circular DNA molecule that functions as a self-replicating genetic element (replicon). Extrachromosomal genetic elements, such as plasmids and bacteriophages, are nonessential replicons that often determine resistance to antimicrobial agents, the production of virulence factors, or other functions. The chromosome replicates semi conservatively; each strand of DNA serves as a template for the synthesis of its complementary strand.
Mutation and Selection
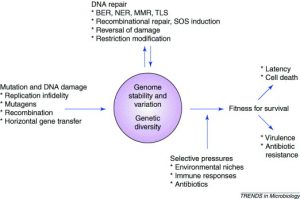
The complete set of genetic determinants of an organism constitutes its genotype, and the observable characteristics constitute its phenotype. Mutations are hereditary changes in the genotype that can occur spontaneously or be induced by chemical or physical treatments. Organisms selected as reference strains are referred to as wild-type and their progeny with mutations are referred to as mutants. Selective media distinguish between wild-type and mutant strains based on growth; differential media distinguish between them based on other phenotypic properties.
Genetic Information Exchange
Genetic exchanges between bacteria occur by several mechanisms. In transformation, the recipient bacterium takes extracellular DNA from the donor. In transduction, the donor DNA packaged in a bacteriophage infects the recipient bacteria. In conjugation, the donor bacterium transfers DNA to the recipient by mating. Recombination of Microbiology is the rearrangement of the donor and recipient genomes to form new hybrid genomes. Transposons are mobile segments of DNA that move from one place to another within or between genomes.
Recombinant DNA and gene cloning
Gene cloning is the incorporation of a foreign gene into a vector to produce a recombinant DNA molecule that replicates and expresses the foreign gene in a recipient cell. Cloned genes are detected by the phenotypes they determine or by the specific nucleotide sequences they contain. Recombinant DNA and gene cloning are essential tools for research in molecular microbiology and medicine. They have many medical applications, including the development of new vaccines, biological products, diagnostic tests, and therapeutic methods.
Regulation of Gene Expression
Gene expression in microbes is often regulated by intracellular or environmental conditions. Regulation can affect any step in gene expression, including the initiation or termination of transcription, translation, or the activity of gene products. An operon is a set of genes that are transcribed as a single unit and expressed in a coordinated manner. Specific regulation induces or represses a particular gene or operon. The global regulation affects a set of operons, which constitute a regulon. All operons in the regulon are coordinately controlled by the same regulatory mechanism.
Genetic information in microbes
The genetic material of bacteria and plasmids is DNA. Bacterial viruses (bacteriophages or phages) have DNA or RNA as their genetic material. The two essential functions of genetic material are replication and expression. The genetic material must be replicated precisely for the progeny to inherit all the specific genetic determinants (the genotype) of the parent organism. The expression of specific genetic material under a particular set of growth conditions determines the observable characteristics (phenotype) of the organism.
Bacteria have few structural or developmental features that can be easily observed, but they do have a wide range of biochemical capabilities and patterns of susceptibility to antimicrobial agents or bacteriophages. These latter characteristics are often selected as inherited traits to be analyzed in bacterial genetics studies. Current status in the expression of recombinant proteins in microbial systems
Without a doubt, E. coli is the most widely used host for the expression of heterologous genes. It has been used for this purpose for more than 40 years, so there is a lot of knowledge accumulated about its advantages and disadvantages as a platform for expression. Rosano and Ceccarelli review the tools available (expression vectors, strains, media composition, etc.) when using E. coli as a host (Rosano and Ceccarelli, 2014). Different approaches to solving common problems, such as inclusion body (IB) formation or poor performance, are also presented.
Other authors delve a little deeper into these issues. Costa and her co-authors provide a detailed description of the different fusion tags that can be added to the target protein to increase its solubility and/or facilitate its purification from the cellular medium (Costa et al., 2014). In this line, Correa and his co-authors present a very promising approach to easily assess the solubility of a recombinant protein by cloning the corresponding gene in 12 different expression vectors in parallel (Correa et al., 2014).
Although the formation of IBs is mainly considered as a nuance in the production of recombinant proteins, Ramon et al argue that this is not always true and focus on the positive side of IBs, highlighting the advantages of producing recombinant proteins such as IBs for basic proteins and applied research (Ramón et al., 2014). The null or low yield may be the result of codon bias and Elena and co-authors discuss one of the strategies used to solve this problem, codon-optimized gene expression (Elena et al., 2014).
Of great importance is his account of the application of this technology in the industrial environment. Not always the desired product is a protein, sometimes the goal is metabolites and fine chemistry. Ceccoli and co-authors’ review details with numerous examples of how E. coli and other microorganisms can become biocatalysts through strain engineering (Ceccoli et al., 2014). In this way, high-value products can be obtained from pure recombinant enzymes or from whole-cell systems, turning the host into an exquisitely designed and environmentally friendly chemical factory.
Molecular Cloning using Transformation
The most widely used mechanism for introducing modified plasmids into a bacterial cell is transformation, a process in which bacteria take up free DNA from their environment. In nature, free DNA normally comes from other lysed bacterial cells; In the laboratory, free DNA in the form of recombinant plasmids is introduced into the cell’s environment.
Some bacteria, such as Bacillus spp., are naturally competent, meaning they can take up foreign DNA. However, not all bacteria are naturally competent. In most cases, bacteria must be rendered artificially competent in the laboratory by increasing the permeability of the cell membrane. This can be achieved through chemical treatments that neutralize charges on the cell membrane or by exposing bacteria to an electrical field that creates microscopic pores in the cell membrane. These methods produce chemically competent or electrocompetent bacteria, respectively.
Following the transformation protocol, the bacterial cells were seeded in an antibiotic-containing medium to inhibit the growth of many host cells that were not transformed by the plasmid conferring antibiotic resistance. Next, a technique called blue-white screening is used for plasmid vectors encoding lacZ, such as pUC19. Blue colonies have a functional beta-galactosidase enzyme because the lacZ gene is not disrupted, with no foreign DNA inserted at the polylinker site.
These colonies typically result from the digested linearized plasmid reattaching to itself. However, the white colonies lack a functional beta-galactosidase enzyme, indicating the insertion of foreign DNA into the polylinker site of the plasmid vector, thus disrupting the lacZ gene. Therefore, the white colonies resulting from this blue-white analysis contain plasmids with an insert and can be further analyzed to characterize foreign DNA. To ensure that the correct DNA was incorporated into the plasmid, the DNA insert can be sequenced.
Molecular cloning by conjugation or transduction
The bacterial process of conjugation (see How Asexual Prokaryotes Achieve Genetic Diversity) can also be manipulated for molecular cloning. F-plasmids, or fertility plasmids, are transferred between bacterial cells through the conjugation process. Recombinant DNA can be transferred by conjugation when bacterial cells containing a recombinant F plasmid are mixed with compatible bacterial cells lacking the plasmid.
F plasmids encode a surface structure called an F pilus that facilitates contact between a cell containing an F plasmid and one without an F plasmid. Upon contact, a cytoplasmic bridge forms between the two cells and the cell containing the F plasmid replicates. its plasmid, by transferring a copy of the recombinant F plasmid to the recipient cell. Once it has received the recombinant F plasmid, the recipient cell can produce its own F pilus and facilitate the transfer of the recombinant F plasmid to an additional cell.
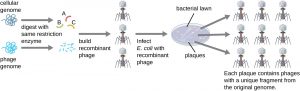
The use of conjugation to transfer recombinant F plasmids into recipient cells is another efficient way of introducing recombinant DNA molecules into host cells. Alternatively, bacteriophages can be used to introduce recombinant DNA into host bacterial cells through manipulation of the transduction process (see How asexual prokaryotes achieve genetic diversity).
In the laboratory, the DNA fragments of interest can be transformed into phagemids, which are plasmids that have phage sequences that allow them to be packaged into bacteriophages. Bacterial cells can then be infected with these bacteriophages so that the recombinant phagemids can be introduced into the bacterial cells. Depending on the type of phage, the recombinant DNA may integrate into the host bacterial genome (lysogeny) or may exist as a plasmid in the host cytoplasm.